Summary
These analyses were published as “Relationship between insomnia and depression in a community sample depends on habitual sleep duration” in the journal Sleep and Biological Rhythms. They were inspired by a study by Perlis et al. using canonical correlations to relate individual sleep EEG features to specific depressive symptoms. The principal findings were that 1) insomnia severity and sleep duration were both associated with individual depression symptoms, 2) insomnia, but not sleep duration, was related to depression severity among depressed individuals (PHQ-9 score > 10), and 3) the interaction between insomnia and short sleep duration in predicting depression severity was small and negative, suggesting shared variance between insomnia and sleep duration.
Dataset
The data used for these analyses derive from the Sleep and Healthy Activity, Diet, Environment, and Socialization (SHADES) Study conducted by Michael Grandner and funded by the National Institute of Environmental Health Sciences (R21ES022931).
Variables
Insomnia severity was measured using the Insomnia Severity Index. Depression severity and symptoms were measured using the PHQ-9. Symptoms were measured individually and combined into an adjusted PHQ-9 total score (excluding the sleep item to avoid collinearity). Sleep duration was derived from a single self-report question used in the NHANES and categorized as Short Sleep (< 7 hours), Recommended Sleep (7-8 hours), and Long Sleep (> 8 hours). Covariates were age, sex, race, education, and BMI (by self-report).
#### Load Packages ####
library(gtsummary)
library(gt)
library(nnet)
library(foreign)
library(ggpubr)
library(car)
library(broom)
library(tidyverse)
#### Load Data ####
phq9.data <- read.dta("c:/users/atala/sync/research/data/shades/shades.dta") %>% as_tibble() %>%
transmute(
"ID" = response_id,
"ISItotal" = isi,
"ISIfalling" = isi1_falling,
"ISIstaying" = isi2_staying,
"ISIema" = isi3_ema,
"ISIsatisfied" = as.numeric(isi4_satisfied) %>%
factor(., levels = c("1","2","3","4","5"),labels = c("Very Satisfied","Satisfied","Neutral","Dissatisfied","Very Dissatisfied")),
"ISIinterfere" = isi5_interfere,
"ISInoticeable" = isi6_noticeable,
"ISIworried" = isi7_worried,
"ISIscore" = case_when(ISItotal < 8 ~ "No Insomnia",
ISItotal %in% 8:14 ~ "Subthreshold Insomnia",
ISItotal %in% 15:21 ~ "Moderate Insomnia",
ISItotal > 21 ~ "Severe Insomnia") %>%
factor(levels=c("No Insomnia","Subthreshold Insomnia","Moderate Insomnia","Severe Insomnia")),
"PHQinterest" = phq1_interest,
"PHQdepress" = phq2_depress,
"PHQsleep" = phq3_sleep,
"PHQtired" = phq4_tired,
"PHQappetite" = phq5_appetite,
"PHQfailure" = phq6_failure,
"PHQconcentrate" = phq7_concentrate,
"PHQpsychomotor" = phq8_psychomotor,
"PHQsuicide" = phq9_suicide,
"PHQdifficulty" = phq10_difficulty,
"PHQtotal" = phq_total,
"PHQtotal2" = PHQtotal - PHQsleep,
"PHQscore" = case_when(PHQtotal < 5 ~ "Minimal Depression",
PHQtotal %in% 5:9 ~ "Mild Depression",
PHQtotal %in% 10:14 ~ "Moderate Depression",
PHQtotal %in% 15:19 ~ "Moderately-Severe Depression",
PHQtotal > 19 ~ "Severe Depression") %>%
factor(levels=c("Minimal Depression","Mild Depression","Moderate Depression",
"Moderately-Severe Depression","Severe Depression")),
"Depressed" = case_when(PHQtotal > 9 ~ "Depressed",
PHQtotal < 10 ~ "Not Depressed") %>%
factor(levels = c("Not Depressed","Depressed")),
"TST" = nhanes_tst,
"SleepDuration" = case_when(TST>8~"Long Sleep",
TST<7~"Short Sleep",
TST %in% 7:8~"Recommended Sleep") %>%
factor(levels = c("Recommended Sleep","Short Sleep","Long Sleep")),
"Age" = demo_age_years2,
"Sex" = demo_sex %>% factor(0:1,c("Male","Female")),
"Race" = case_when(demo_race_white_only==1~"White",
demo_race_black_only==1~"Black",
demo_race_asian_only==1~"Asian",
demo_race_hisp_only==1~"Hispanic",
demo_race_native_only==1~"Native",
demo_race_pacisland_only==1~"Other",
demo_race_multiracial==1~"Other") %>%
factor(levels = c("White","Black","Asian","Hispanic","Native","Other")) %>%
replace_na(., "Other"),
"Education" = factor(demo_education2, 0:3,c("College","Some College","High School","Less than high school")),
"BMI" = body_weight_curr_kg/((body_height_cm)/100)^2
)
Table 1
The characteristics of the sample are presented below in Table 1.
#### Table 1: Demographics and Response Characteristics of the Sample ####
tbl_summary(
phq9.data,
label = list("Age" = "Age (years)", "BMI" = "BMI (kg/m^2)", "ISIscore" = "ISI Category","PHQscore" = "PHQ-9 Category"),
by="SleepDuration",
include = c("SleepDuration","Age","BMI","Sex","Education","Race","ISIscore","PHQscore"),
statistic = list(all_continuous() ~ "{mean} ({sd})", all_categorical() ~ "{n} ({p}%)"),
digits = list(all_continuous() ~ c(1,2), all_categorical() ~ c(0,1))
) %>% add_overall() %>% add_p(test = list(all_continuous() ~ "aov", all_categorical() ~ "chisq.test"))
| Characteristic | Overall, N = 1,0071 | Recommended Sleep, N = 4801 | Short Sleep, N = 4771 | Long Sleep, N = 501 | p-value2 |
|---|---|---|---|---|---|
| Age (years) | 34.0 (9.45) | 32.8 (9.33) | 35.6 (9.54) | 30.2 (6.51) | <0.001 |
| BMI (kg/m^2) | 26.6 (7.85) | 25.3 (5.63) | 27.9 (9.19) | 27.4 (10.01) | <0.001 |
| Sex | 0.082 | ||||
| Male | 388 (38.5%) | 169 (35.2%) | 201 (42.1%) | 18 (36.0%) | |
| Female | 619 (61.5%) | 311 (64.8%) | 276 (57.9%) | 32 (64.0%) | |
| Education | <0.001 | ||||
| College | 563 (55.9%) | 323 (67.3%) | 205 (43.0%) | 35 (70.0%) | |
| Some College | 312 (31.0%) | 118 (24.6%) | 181 (37.9%) | 13 (26.0%) | |
| High School | 106 (10.5%) | 31 (6.5%) | 73 (15.3%) | 2 (4.0%) | |
| Less than high school | 26 (2.6%) | 8 (1.7%) | 18 (3.8%) | 0 (0.0%) | |
| Race | <0.001 | ||||
| White | 597 (59.3%) | 324 (67.5%) | 242 (50.7%) | 31 (62.0%) | |
| Black | 251 (24.9%) | 83 (17.3%) | 162 (34.0%) | 6 (12.0%) | |
| Asian | 55 (5.5%) | 32 (6.7%) | 23 (4.8%) | 0 (0.0%) | |
| Hispanic | 46 (4.6%) | 17 (3.5%) | 23 (4.8%) | 6 (12.0%) | |
| Native | 3 (0.3%) | 1 (0.2%) | 2 (0.4%) | 0 (0.0%) | |
| Other | 55 (5.5%) | 23 (4.8%) | 25 (5.2%) | 7 (14.0%) | |
| ISI Category | <0.001 | ||||
| No Insomnia | 350 (34.8%) | 252 (52.5%) | 78 (16.4%) | 20 (40.0%) | |
| Subthreshold Insomnia | 389 (38.6%) | 171 (35.6%) | 199 (41.7%) | 19 (38.0%) | |
| Moderate Insomnia | 212 (21.1%) | 55 (11.5%) | 146 (30.6%) | 11 (22.0%) | |
| Severe Insomnia | 56 (5.6%) | 2 (0.4%) | 54 (11.3%) | 0 (0.0%) | |
| PHQ-9 Category | <0.001 | ||||
| Minimal Depression | 308 (30.6%) | 201 (41.9%) | 94 (19.7%) | 13 (26.0%) | |
| Mild Depression | 309 (30.7%) | 154 (32.1%) | 138 (28.9%) | 17 (34.0%) | |
| Moderate Depression | 209 (20.8%) | 78 (16.2%) | 123 (25.8%) | 8 (16.0%) | |
| Moderately-Severe Depression | 108 (10.7%) | 28 (5.8%) | 74 (15.5%) | 6 (12.0%) | |
| Severe Depression | 73 (7.2%) | 19 (4.0%) | 48 (10.1%) | 6 (12.0%) | |
| 1 Mean (SD); n (%) | |||||
| 2 One-way ANOVA; Pearson's Chi-squared test | |||||
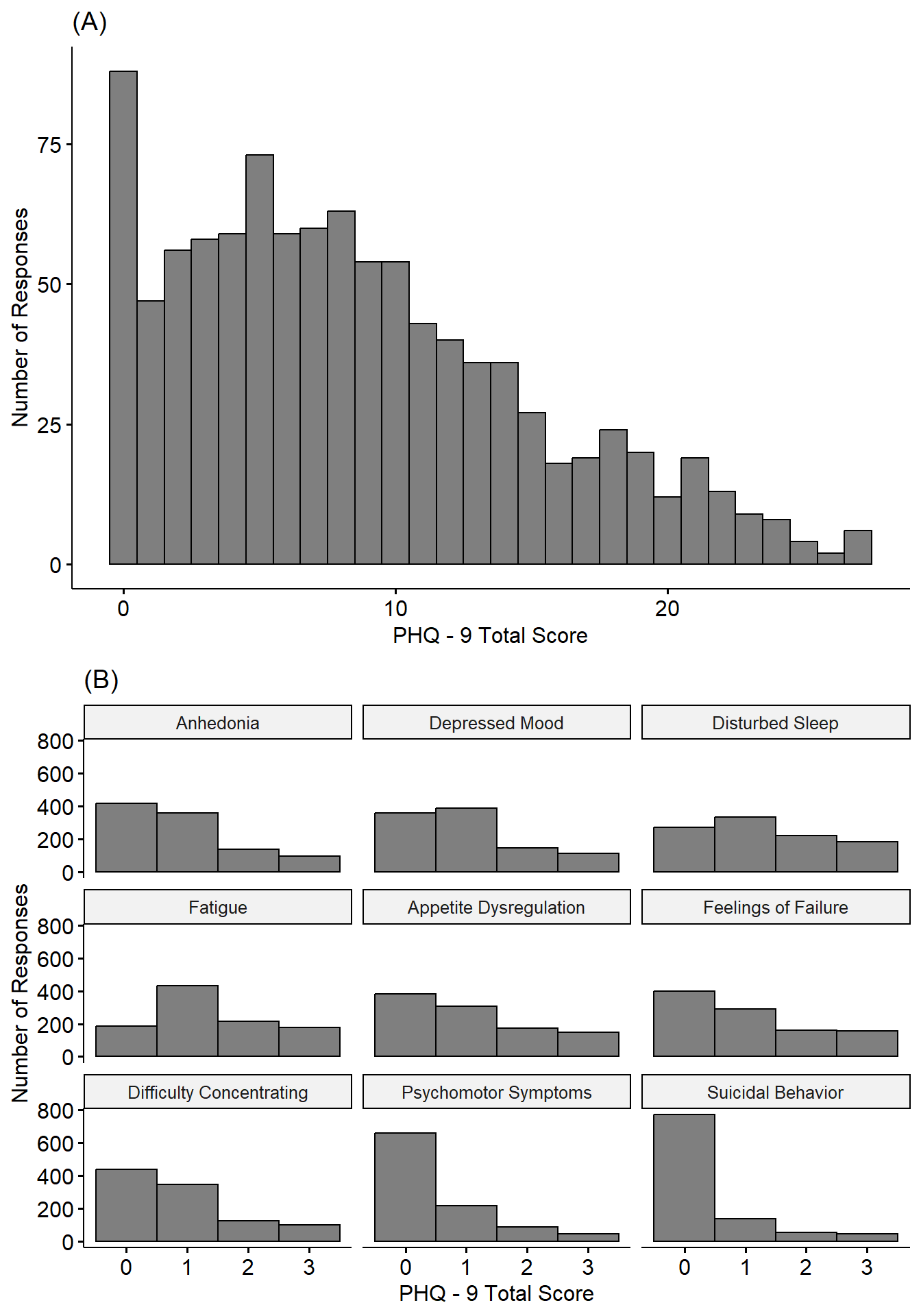
Data Distribution
Let’s look at the distribution of the data.
#### Histograms to justify QP modeling ####
phq9.hist <- phq9.data %>% select(ID,PHQinterest,PHQdepress,PHQsleep,PHQtired,PHQappetite,
PHQfailure,PHQconcentrate,PHQpsychomotor,PHQsuicide) %>%
gather("Symptom","Frequency",-"ID") %>%
mutate(Symptom = factor(Symptom,levels=c("PHQinterest","PHQdepress","PHQsleep","PHQtired","PHQappetite",
"PHQfailure","PHQconcentrate","PHQpsychomotor","PHQsuicide"),
labels = c("Anhedonia","Depressed Mood","Disturbed Sleep","Fatigue","Appetite Dysregulation",
"Feelings of Failure","Difficulty Concentrating","Psychomotor Symptoms","Suicidal Behavior"))
)
totalphq.plot <- gghistogram(phq9.data,x="PHQtotal",legend = "none",ylab = "Number of Responses",xlab = "PHQ - 9 Total Score",
title = "(A)",bins=28,ggtheme=theme_pubr(),fill="black")
itemphq.plot <- gghistogram(phq9.hist,x="Frequency",legend = "none",ylab = "Number of Responses",xlab = "PHQ - 9 Total Score",
title = "(B)",bins=28,facet.by = "Symptom",ggtheme=theme_pubr(),fill="black",binwidth = 1)
The simplest approach to relate insomnia and sleep duration to depression would be using linear regression. This assumes, however, that the outcome (i.e., depression severity and frequency of symptoms) are normally distributed. Using histograms, however, it is clear these data are not normally distributed. In fact, they follow a Poisson-like distribution, with most responses skewed toward smaller values.
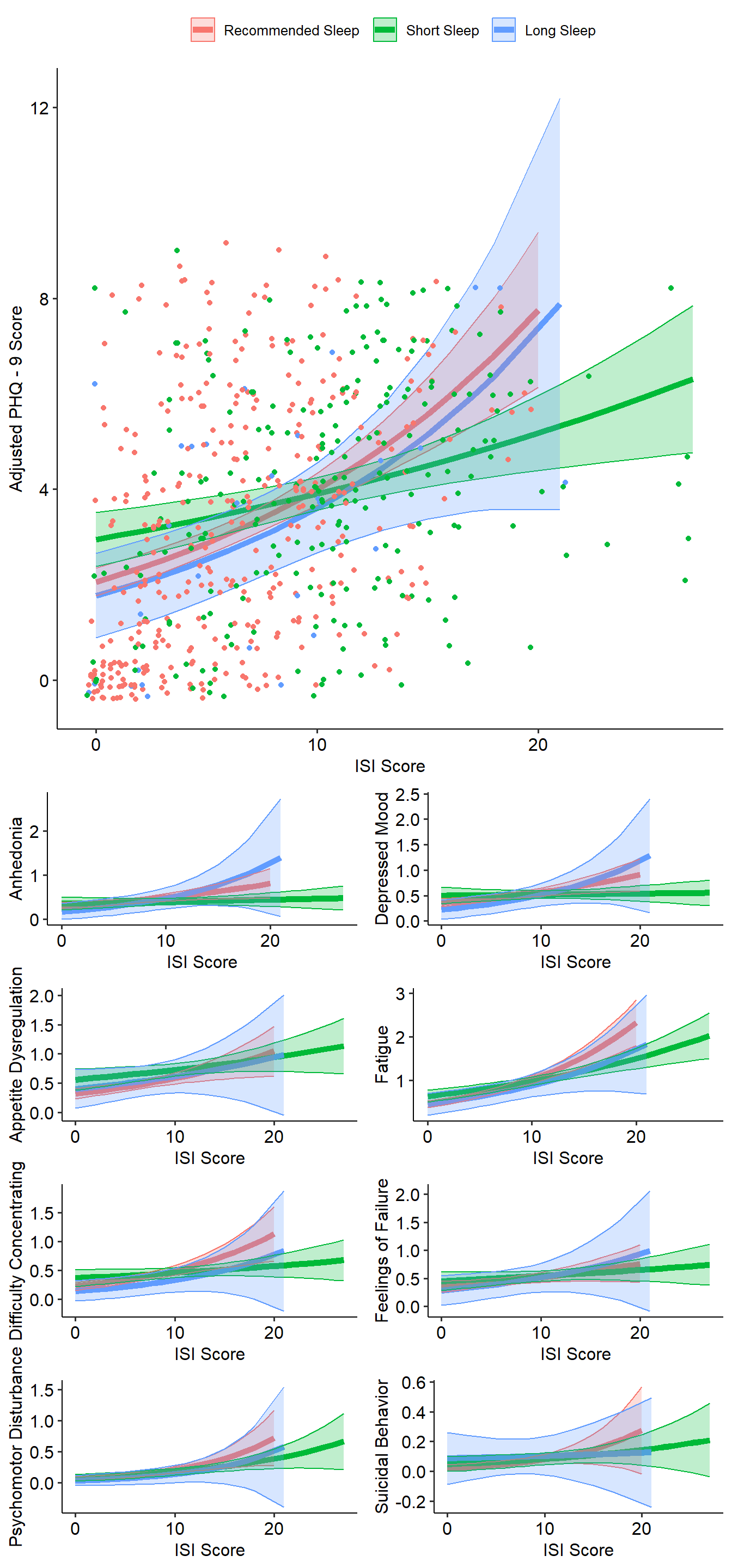
Model the Whole Sample
The following code models the adjusted PHQ-9 score (“PHQtotal2”) as a function of insomnia severity and sleep duration. Models are unadjusted and adjusted for age, sex, race, and ethnicity.
#### Analysis 1) Model adjPHQ-9 Score as a function of ISI score and sleep duration in the whole sample ####
## Total Score
# Unadjusted
phqscore.qp <- glm(PHQtotal2~ISItotal*SleepDuration,family="quasipoisson",phq9.data)
phqscore.qp.maineff <- bind_cols("Symptom" = rep("Total",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(phqscore.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(phqscore.qp)$coefficients[2:6,4])
# Adjusted for Age, Sex, Race, Education, BMI
phqscore.qp.full <- glm(PHQtotal2~ISItotal*SleepDuration+Age+Sex+Race+Education+BMI,family="quasipoisson",phq9.data)
phqscore.qp.full.maineff <- bind_cols("Symptom" = rep("Total",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(phqscore.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(phqscore.qp.full)$coefficients[c(2:4,16,17),4])
# Plot PHQScore vs. ISItotal by SleepDuration
phqscore.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQtotal2),
predict(phqscore.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
geom_point(aes(y = PHQtotal2),position="jitter")+
labs(x = "ISI Score",y="Adjusted PHQ - 9 Score")+
theme_pubr()+theme(legend.title = element_blank())
## Individual Symptoms
# Anhedonia
interest.qp <- glm(PHQinterest~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
interest.qp.maineff <- bind_cols("Symptom" = rep("Interest",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(interest.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(interest.qp)$coefficients[2:6,4])
interest.qp.full <- glm(PHQinterest~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
interest.qp.full.maineff <- bind_cols("Symptom" = rep("Interest",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(interest.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(interest.qp.full)$coefficients[c(2:4,16,17),4])
interest.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQinterest),
predict(interest.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Anhedonia")+
theme_pubr()+theme(legend.position = "none")
# Depressed Mood
depress.qp <- glm(PHQdepress~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
depress.qp.maineff <- bind_cols("Symptom" = rep("Depress",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(depress.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(depress.qp)$coefficients[2:6,4])
depress.qp.full <- glm(PHQdepress~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
depress.qp.full.maineff <- bind_cols("Symptom" = rep("Depress",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(depress.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(depress.qp.full)$coefficients[c(2:4,16,17),4])
depress.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQdepress),
predict(depress.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Depressed Mood")+
theme_pubr()+theme(legend.position = "none")
# Appetite
appetite.qp <- glm(PHQappetite~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
appetite.qp.maineff <- bind_cols("Symptom" = rep("Appetite",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(appetite.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(appetite.qp)$coefficients[2:6,4])
appetite.qp.full <- glm(PHQappetite~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
appetite.qp.full.maineff <- bind_cols("Symptom" = rep("Appetite",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(appetite.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(appetite.qp.full)$coefficients[c(2:4,16,17),4])
appetite.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQappetite),
predict(appetite.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Appetite Dysregulation")+
theme_pubr()+theme(legend.position = "none")
# Fatigue
tired.qp <- glm(PHQtired~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
tired.qp.maineff <- bind_cols("Symptom" = rep("Tired",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(tired.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(tired.qp)$coefficients[2:6,4])
tired.qp.full <- glm(PHQtired~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
tired.qp.full.maineff <- bind_cols("Symptom" = rep("Tired",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(tired.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(tired.qp.full)$coefficients[c(2:4,16,17),4])
tired.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQtired),
predict(tired.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Fatigue")+
theme_pubr()+theme(legend.position = "none")
# Difficulty Concentrating
concentrate.qp <- glm(PHQconcentrate~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
concentrate.qp.maineff <- bind_cols("Symptom" = rep("Concentrate",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(concentrate.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(concentrate.qp)$coefficients[2:6,4])
concentrate.qp.full <- glm(PHQconcentrate~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
concentrate.qp.full.maineff <- bind_cols("Symptom" = rep("Concentrate",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(concentrate.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(concentrate.qp.full)$coefficients[c(2:4,16,17),4])
concentrate.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQconcentrate),
predict(concentrate.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Difficulty Concentrating")+
theme_pubr()+theme(legend.position = "none")
# Feelings of Failure
failure.qp <- glm(PHQfailure~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
failure.qp.maineff <- bind_cols("Symptom" = rep("Failure",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(failure.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(failure.qp)$coefficients[2:6,4])
failure.qp.full <- glm(PHQfailure~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
failure.qp.full.maineff <- bind_cols("Symptom" = rep("Failure",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(failure.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(failure.qp.full)$coefficients[c(2:4,16,17),4])
failure.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQfailure),
predict(failure.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Feelings of Failure")+
theme_pubr()+theme(legend.position = "none")
# Psychomotor Disturbance
psychomotor.qp <- glm(PHQpsychomotor~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
psychomotor.qp.maineff <- bind_cols("Symptom" = rep("Psychomotor",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(psychomotor.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(psychomotor.qp)$coefficients[2:6,4])
psychomotor.qp.full <- glm(PHQpsychomotor~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
psychomotor.qp.full.maineff <- bind_cols("Symptom" = rep("Psychomotor",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(psychomotor.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(psychomotor.qp.full)$coefficients[c(2:4,16,17),4])
psychomotor.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQpsychomotor),
predict(psychomotor.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Psychomotor Disturbance")+
theme_pubr()+theme(legend.position = "none")
# Suicidal behavior
suicide.qp <- glm(PHQsuicide~ISItotal*SleepDuration,phq9.data,family="quasipoisson")
suicide.qp.maineff <- bind_cols("Symptom" = rep("Suicide",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(suicide.qp, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(suicide.qp)$coefficients[2:6,4])
suicide.qp.full <- glm(PHQsuicide~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.data,family="quasipoisson")
suicide.qp.full.maineff <- bind_cols("Symptom" = rep("Suicide",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(suicide.qp.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(suicide.qp.full)$coefficients[c(2:4,16,17),4])
suicide.plot <- ggplot(
data = bind_cols(phq9.data %>% select(ISItotal,SleepDuration,PHQsuicide),
predict(suicide.qp, type="response",se.fit=TRUE)),
aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,
fill=SleepDuration),
alpha=.25)+
labs(x = "ISI Score",y="Suicidal Behavior")+
theme_pubr()+theme(legend.position = "none")
#### Table 2: Sleep influence by symptom in unadjusted and adjusted models ####
phq9.symptom.summary <- bind_cols(
bind_rows(phqscore.qp.maineff,interest.qp.maineff,depress.qp.maineff,appetite.qp.maineff,tired.qp.maineff,
concentrate.qp.maineff,failure.qp.maineff,psychomotor.qp.maineff,suicide.qp.maineff) %>%
transmute(Symptom = factor(Symptom,levels = c("Total","Interest","Depress","Appetite","Tired","Concentrate","Failure","Psychomotor","Suicide"),
labels = c("Adjusted PHQ-9 Score","Anhedonia","Depressed Mood","Appetite dysregulation","Fatigue",
"Difficulty concentrating","Feelings of failure","Psychomotor disturbance","Suicidal behavior")),
Parameter,
"PR" = round(Estimate,2),
"SE" = round(SE,2),
"95% CI" = paste0("[",round(`2.5 %`,2),", ",round(`97.5 %`,2),"]"),
"p-value" = round(`p-value`,4)),
bind_rows(phqscore.qp.full.maineff,interest.qp.full.maineff,depress.qp.full.maineff,appetite.qp.full.maineff,tired.qp.full.maineff,
concentrate.qp.full.maineff,failure.qp.full.maineff,psychomotor.qp.full.maineff,suicide.qp.full.maineff) %>%
transmute("aPR" = round(Estimate,2),
"aSE" = round(SE,2),
"a95% CI" = paste0("[",round(`2.5 %`,2),", ",round(`97.5 %`,2),"]"),
"ap-value" = round(`p-value`,4))
)
The results of these models are presented in the table below. The predictors of interest are ISI score, sleep duration, and their interaction. These are reported for each symptom and the overall PHQ-9 severity score (minus the sleep item due to collinearity). The adjusted model results are in the right half of the table with headers appended by “a”.
| Symptom | Parameter | PR | SE | 95% CI | p-value | aPR | aSE | a95% CI | ap-value |
|---|---|---|---|---|---|---|---|---|---|
| Adjusted PHQ-9 Score | ISI | 1.09 | 0.01 | [1.08, 1.11] | 0.0000 | 1.09 | 0.01 | [1.08, 1.1] | 0.0000 |
| Adjusted PHQ-9 Score | Short Sleep | 1.52 | 0.16 | [1.21, 1.83] | 0.0001 | 1.48 | 0.15 | [1.18, 1.78] | 0.0002 |
| Adjusted PHQ-9 Score | Long Sleep | 1.40 | 0.31 | [0.8, 2] | 0.1247 | 1.35 | 0.30 | [0.77, 1.93] | 0.1708 |
| Adjusted PHQ-9 Score | ISI:Short Sleep | 0.97 | 0.01 | [0.95, 0.98] | 0.0000 | 0.97 | 0.01 | [0.96, 0.99] | 0.0001 |
| Adjusted PHQ-9 Score | ISI:Long Sleep | 0.99 | 0.02 | [0.95, 1.02] | 0.4114 | 0.99 | 0.02 | [0.95, 1.02] | 0.4774 |
| Anhedonia | ISI | 1.08 | 0.01 | [1.06, 1.1] | 0.0000 | 1.08 | 0.01 | [1.06, 1.1] | 0.0000 |
| Anhedonia | Short Sleep | 1.08 | 0.17 | [0.76, 1.41] | 0.6076 | 1.07 | 0.17 | [0.74, 1.39] | 0.6766 |
| Anhedonia | Long Sleep | 1.16 | 0.37 | [0.44, 1.88] | 0.6347 | 1.15 | 0.37 | [0.43, 1.86] | 0.6710 |
| Anhedonia | ISI:Short Sleep | 0.99 | 0.01 | [0.97, 1.01] | 0.2291 | 0.99 | 0.01 | [0.97, 1.01] | 0.2924 |
| Anhedonia | ISI:Long Sleep | 1.01 | 0.02 | [0.96, 1.06] | 0.6572 | 1.01 | 0.02 | [0.96, 1.06] | 0.6143 |
| Depressed Mood | ISI | 1.09 | 0.01 | [1.07, 1.1] | 0.0000 | 1.08 | 0.01 | [1.07, 1.1] | 0.0000 |
| Depressed Mood | Short Sleep | 1.32 | 0.18 | [0.96, 1.68] | 0.0430 | 1.29 | 0.18 | [0.94, 1.64] | 0.0699 |
| Depressed Mood | Long Sleep | 1.32 | 0.38 | [0.57, 2.07] | 0.3342 | 1.31 | 0.38 | [0.56, 2.06] | 0.3546 |
| Depressed Mood | ISI:Short Sleep | 0.97 | 0.01 | [0.95, 0.99] | 0.0033 | 0.97 | 0.01 | [0.95, 0.99] | 0.0096 |
| Depressed Mood | ISI:Long Sleep | 0.99 | 0.02 | [0.95, 1.03] | 0.6561 | 0.99 | 0.02 | [0.95, 1.03] | 0.6471 |
| Appetite dysregulation | ISI | 1.09 | 0.01 | [1.07, 1.11] | 0.0000 | 1.09 | 0.01 | [1.07, 1.1] | 0.0000 |
| Appetite dysregulation | Short Sleep | 1.86 | 0.26 | [1.34, 2.38] | 0.0000 | 1.82 | 0.26 | [1.31, 2.33] | 0.0000 |
| Appetite dysregulation | Long Sleep | 1.89 | 0.55 | [0.81, 2.97] | 0.0291 | 1.76 | 0.51 | [0.76, 2.76] | 0.0508 |
| Appetite dysregulation | ISI:Short Sleep | 0.96 | 0.01 | [0.94, 0.98] | 0.0004 | 0.97 | 0.01 | [0.95, 0.99] | 0.0009 |
| Appetite dysregulation | ISI:Long Sleep | 0.96 | 0.02 | [0.92, 1.01] | 0.1135 | 0.97 | 0.02 | [0.92, 1.01] | 0.1486 |
| Fatigue | ISI | 1.08 | 0.01 | [1.07, 1.09] | 0.0000 | 1.08 | 0.01 | [1.07, 1.09] | 0.0000 |
| Fatigue | Short Sleep | 1.51 | 0.14 | [1.22, 1.79] | 0.0000 | 1.49 | 0.14 | [1.21, 1.77] | 0.0000 |
| Fatigue | Long Sleep | 1.16 | 0.25 | [0.68, 1.65] | 0.4775 | 1.13 | 0.24 | [0.66, 1.6] | 0.5616 |
| Fatigue | ISI:Short Sleep | 0.97 | 0.01 | [0.95, 0.98] | 0.0000 | 0.97 | 0.01 | [0.96, 0.99] | 0.0001 |
| Fatigue | ISI:Long Sleep | 0.99 | 0.02 | [0.96, 1.03] | 0.6999 | 0.99 | 0.02 | [0.96, 1.03] | 0.7458 |
| Difficulty concentrating | ISI | 1.10 | 0.01 | [1.08, 1.12] | 0.0000 | 1.09 | 0.01 | [1.07, 1.11] | 0.0000 |
| Difficulty concentrating | Short Sleep | 1.61 | 0.26 | [1.1, 2.12] | 0.0031 | 1.58 | 0.26 | [1.08, 2.09] | 0.0049 |
| Difficulty concentrating | Long Sleep | 1.23 | 0.45 | [0.34, 2.12] | 0.5797 | 1.13 | 0.42 | [0.31, 1.94] | 0.7432 |
| Difficulty concentrating | ISI:Short Sleep | 0.96 | 0.01 | [0.94, 0.99] | 0.0013 | 0.97 | 0.01 | [0.95, 0.99] | 0.0064 |
| Difficulty concentrating | ISI:Long Sleep | 0.98 | 0.03 | [0.93, 1.04] | 0.5120 | 0.98 | 0.03 | [0.93, 1.04] | 0.5859 |
| Feelings of failure | ISI | 1.08 | 0.01 | [1.07, 1.1] | 0.0000 | 1.08 | 0.01 | [1.06, 1.1] | 0.0000 |
| Feelings of failure | Short Sleep | 1.58 | 0.23 | [1.12, 2.04] | 0.0020 | 1.54 | 0.23 | [1.09, 1.99] | 0.0040 |
| Feelings of failure | Long Sleep | 1.45 | 0.44 | [0.58, 2.32] | 0.2256 | 1.43 | 0.44 | [0.57, 2.3] | 0.2404 |
| Feelings of failure | ISI:Short Sleep | 0.97 | 0.01 | [0.95, 0.99] | 0.0018 | 0.97 | 0.01 | [0.95, 0.99] | 0.0047 |
| Feelings of failure | ISI:Long Sleep | 0.99 | 0.02 | [0.94, 1.04] | 0.6448 | 0.99 | 0.02 | [0.94, 1.04] | 0.6494 |
| Psychomotor disturbance | ISI | 1.13 | 0.02 | [1.1, 1.17] | 0.0000 | 1.12 | 0.02 | [1.09, 1.15] | 0.0000 |
| Psychomotor disturbance | Short Sleep | 1.74 | 0.44 | [0.87, 2.61] | 0.0298 | 1.50 | 0.38 | [0.76, 2.24] | 0.1078 |
| Psychomotor disturbance | Long Sleep | 1.65 | 0.94 | [-0.2, 3.5] | 0.3821 | 1.46 | 0.83 | [-0.16, 3.08] | 0.5075 |
| Psychomotor disturbance | ISI:Short Sleep | 0.96 | 0.02 | [0.93, 0.99] | 0.0176 | 0.97 | 0.02 | [0.94, 1] | 0.0761 |
| Psychomotor disturbance | ISI:Long Sleep | 0.96 | 0.04 | [0.88, 1.04] | 0.3819 | 0.97 | 0.04 | [0.89, 1.06] | 0.5552 |
| Suicidal behavior | ISI | 1.16 | 0.02 | [1.12, 1.2] | 0.0000 | 1.15 | 0.02 | [1.11, 1.19] | 0.0000 |
| Suicidal behavior | Short Sleep | 2.54 | 0.87 | [0.83, 4.26] | 0.0068 | 2.38 | 0.81 | [0.79, 3.96] | 0.0112 |
| Suicidal behavior | Long Sleep | 2.75 | 1.78 | [-0.74, 6.24] | 0.1193 | 2.75 | 1.80 | [-0.77, 6.27] | 0.1213 |
| Suicidal behavior | ISI:Short Sleep | 0.94 | 0.02 | [0.89, 0.98] | 0.0041 | 0.94 | 0.02 | [0.89, 0.98] | 0.0046 |
| Suicidal behavior | ISI:Long Sleep | 0.96 | 0.05 | [0.87, 1.05] | 0.4221 | 0.97 | 0.05 | [0.87, 1.06] | 0.4555 |
Figure 2 below illustrates how changes in ISI score are related to adjusted PHQ-9 score by sleep duration category.
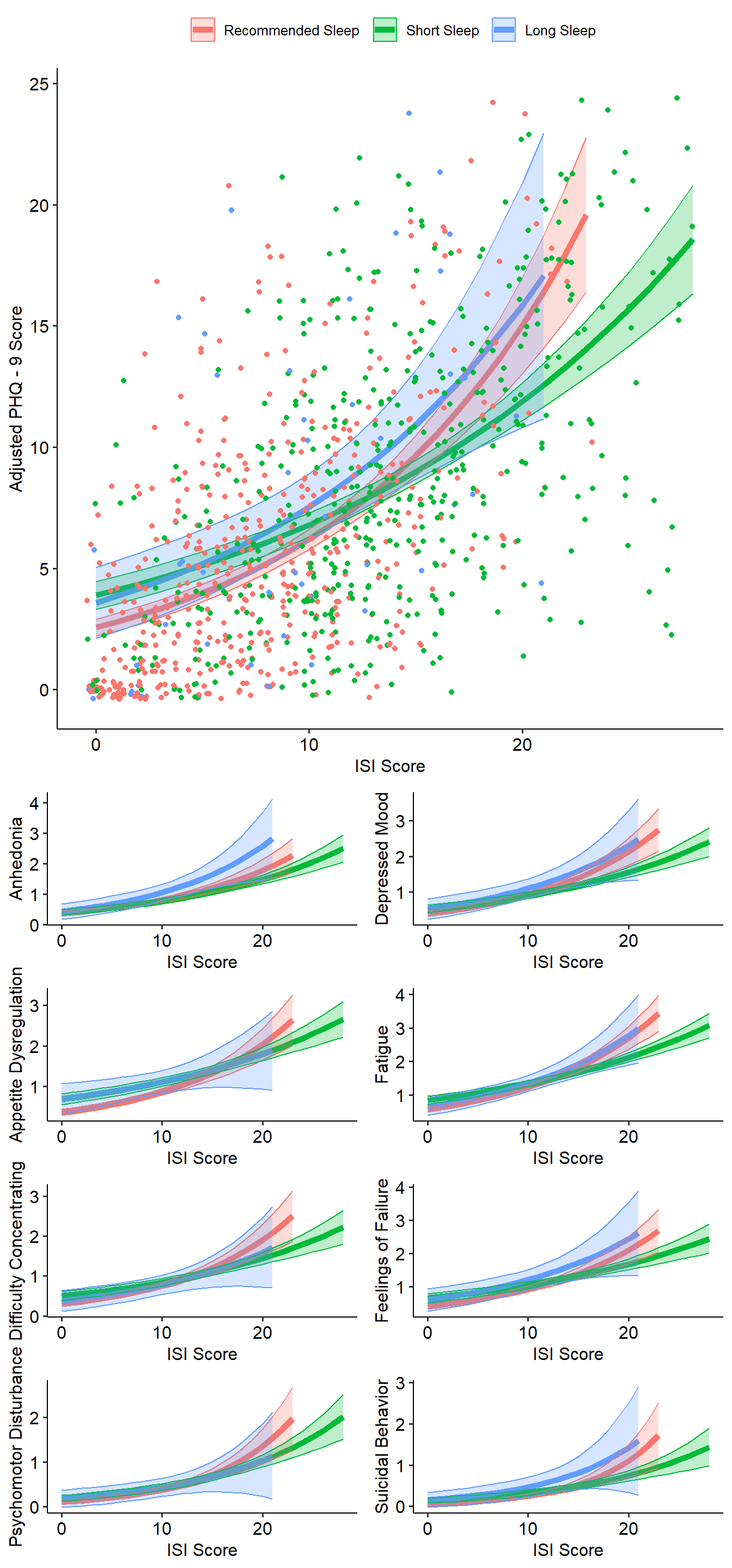
Model Depressed Subjects Only
Let’s look at only the N=390 individuals with clinically significant depression symptoms (PHQ-9 > 10).
#### Analysis 2: Depressed Subjects Only ####
## Total Score
# Unadjusted
phq9.subset <- phq9.data[phq9.data$PHQtotal>9,]
phqscore.qp.sub <- glm(PHQtotal2~ISItotal*SleepDuration,family="quasipoisson",phq9.subset)
phqscore.qp.sub.maineff <- bind_cols("Symptom" = rep("Total",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(phqscore.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(phqscore.qp.sub)$coefficients[2:6,4])
# Adjusted for Age, Sex, Race, Education, and BMI
phqscore.qp.sub.full <- glm(PHQtotal2~ISItotal*SleepDuration+Age+Sex+Race+Education+BMI,family="quasipoisson",phq9.subset)
phqscore.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Total",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(phqscore.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(phqscore.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict <- predict(phqscore.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
phqscore.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
geom_point(aes(y = PHQtotal2),position="jitter")+
labs(x = "ISI Score",y="Adjusted PHQ - 9 Score")+
theme_pubr()+theme(legend.title = element_blank())
## Individual Symptoms
# Anhedonia
interest.qp.sub <- glm(PHQinterest~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
interest.qp.sub.maineff <- bind_cols("Symptom" = rep("Interest",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(interest.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(interest.qp.sub)$coefficients[2:6,4])
interest.qp.sub.full <- glm(PHQinterest~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
interest.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Interest",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(interest.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(interest.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(interest.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
interest.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Anhedonia")+
theme_pubr()+theme(legend.position = "none")
# Depressed Mood
depress.qp.sub <- glm(PHQdepress~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
depress.qp.sub.maineff <- bind_cols("Symptom" = rep("Depress",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(depress.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(depress.qp.sub)$coefficients[2:6,4])
depress.qp.sub.full <- glm(PHQdepress~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
depress.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Depress",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(depress.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(depress.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(depress.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
depress.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Depressed Mood")+
theme_pubr()+theme(legend.position = "none")
# Appetite
appetite.qp.sub <- glm(PHQappetite~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
appetite.qp.sub.maineff <- bind_cols("Symptom" = rep("Appetite",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(appetite.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(appetite.qp.sub)$coefficients[2:6,4])
appetite.qp.sub.full <- glm(PHQappetite~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
appetite.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Appetite",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(appetite.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(appetite.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(appetite.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
appetite.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Appetite Dysregulation")+
theme_pubr()+theme(legend.position = "none")
# Fatigue
tired.qp.sub <- glm(PHQtired~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
tired.qp.sub.maineff <- bind_cols("Symptom" = rep("Tired",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(tired.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(tired.qp.sub)$coefficients[2:6,4])
tired.qp.sub.full <- glm(PHQtired~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
tired.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Tired",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(tired.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(tired.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(tired.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
tired.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Fatigue")+
theme_pubr()+theme(legend.position = "none")
# Difficulty Concentrating
concentrate.qp.sub <- glm(PHQconcentrate~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
concentrate.qp.sub.maineff <- bind_cols("Symptom" = rep("Concentrate",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(concentrate.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(concentrate.qp.sub)$coefficients[2:6,4])
concentrate.qp.sub.full <- glm(PHQconcentrate~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
concentrate.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Concentrate",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(concentrate.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(concentrate.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(concentrate.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
concentrate.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Difficulty Concentrating")+
theme_pubr()+theme(legend.position = "none")
# Feelings of Failure
failure.qp.sub <- glm(PHQfailure~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
failure.qp.sub.maineff <- bind_cols("Symptom" = rep("Failure",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(failure.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(failure.qp.sub)$coefficients[2:6,4])
failure.qp.sub.full <- glm(PHQfailure~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
failure.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Failure",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(failure.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(failure.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(failure.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
failure.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Feelings of Failure")+
theme_pubr()+theme(legend.position = "none")
# Psychomotor Disturbance
psychomotor.qp.sub <- glm(PHQpsychomotor~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
psychomotor.qp.sub.maineff <- bind_cols("Symptom" = rep("Psychomotor",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(psychomotor.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(psychomotor.qp.sub)$coefficients[2:6,4])
psychomotor.qp.sub.full <- glm(PHQpsychomotor~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
psychomotor.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Psychomotor",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(psychomotor.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(psychomotor.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(psychomotor.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
psychomotor.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Psychomotor Disturbance")+
theme_pubr()+theme(legend.position = "none")
# Suicidal behavior
suicide.qp.sub <- glm(PHQsuicide~ISItotal*SleepDuration,phq9.subset,family="quasipoisson")
suicide.qp.sub.maineff <- bind_cols("Symptom" = rep("Suicide",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(suicide.qp.sub, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(suicide.qp.sub)$coefficients[2:6,4])
suicide.qp.sub.full <- glm(PHQsuicide~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset,family="quasipoisson")
suicide.qp.sub.full.maineff <- bind_cols("Symptom" = rep("Suicide",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(suicide.qp.sub.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(suicide.qp.sub.full)$coefficients[c(2:4,16,17),4])
phq9.subset.predict<- predict(suicide.qp.sub, type="response",se.fit=TRUE)
phq9.subset$fit <- as.numeric(phq9.subset.predict$fit)
phq9.subset$se.fit <- as.numeric(phq9.subset.predict$se.fit)
suicide.sub.plot <- ggplot(phq9.subset, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Suicidal Behavior")+
theme_pubr()+theme(legend.position = "none")
#### Table 3: Sleep influence by symptom in unadjusted/adjusted models for depressed only ####
phq9.symptom.sub.summary <- bind_cols(
bind_rows(phqscore.qp.sub.maineff,interest.qp.sub.maineff,depress.qp.sub.maineff,appetite.qp.sub.maineff,tired.qp.sub.maineff,
concentrate.qp.sub.maineff,failure.qp.sub.maineff,psychomotor.qp.sub.maineff,suicide.qp.sub.maineff) %>%
transmute(Symptom = factor(Symptom,levels = c("Total","Interest","Depress","Appetite","Tired","Concentrate","Failure","Psychomotor","Suicide"),
labels = c("Adjusted PHQ-9 Score","Anhedonia","Depressed Mood","Appetite dysregulation","Fatigue",
"Difficulty concentrating","Feelings of failure","Psychomotor disturbance","Suicidal behavior")),
Parameter,
"PR" = Estimate,
"SE" = round(SE,2),
"95% CI" = paste0("[",round(`2.5 %`,2),", ",round(`97.5 %`,2),"]"),
"p-value" = round(`p-value`,4)),
bind_rows(phqscore.qp.sub.full.maineff,interest.qp.sub.full.maineff,depress.qp.sub.full.maineff,appetite.qp.sub.full.maineff,
tired.qp.sub.full.maineff,concentrate.qp.sub.full.maineff,failure.qp.sub.full.maineff,psychomotor.qp.sub.full.maineff,
suicide.qp.sub.full.maineff) %>%
transmute("aPR" = Estimate,
"aSE" = round(SE,2),
"a95% CI" = paste0("[",round(`2.5 %`,2),", ",round(`97.5 %`,2),"]"),
"ap-value" = round(`p-value`,4))
)
The table below shows their model results (as with the whole sample).
gt(phq9.symptom.sub.summary)
| Symptom | Parameter | PR | SE | 95% CI | p-value | aPR | aSE | a95% CI | ap-value |
|---|---|---|---|---|---|---|---|---|---|
| Adjusted PHQ-9 Score | ISI | 1.0178209 | 0.01 | [1.01, 1.03] | 0.0013 | 1.0161896 | 0.01 | [1.01, 1.03] | 0.0038 |
| Adjusted PHQ-9 Score | Short Sleep | 0.9415744 | 0.09 | [0.77, 1.12] | 0.5225 | 0.9304414 | 0.09 | [0.76, 1.1] | 0.4512 |
| Adjusted PHQ-9 Score | Long Sleep | 1.2831456 | 0.25 | [0.79, 1.77] | 0.2002 | 1.2820901 | 0.25 | [0.79, 1.78] | 0.2062 |
| Adjusted PHQ-9 Score | ISI:Short Sleep | 1.0023778 | 0.01 | [0.99, 1.02] | 0.7119 | 1.0038357 | 0.01 | [0.99, 1.02] | 0.5588 |
| Adjusted PHQ-9 Score | ISI:Long Sleep | 0.9917789 | 0.01 | [0.96, 1.02] | 0.5808 | 0.9918640 | 0.02 | [0.96, 1.02] | 0.5895 |
| Anhedonia | ISI | 1.0035278 | 0.01 | [0.98, 1.02] | 0.7128 | 1.0045149 | 0.01 | [0.99, 1.02] | 0.6453 |
| Anhedonia | Short Sleep | 0.6228047 | 0.10 | [0.42, 0.83] | 0.0046 | 0.6395063 | 0.11 | [0.43, 0.85] | 0.0090 |
| Anhedonia | Long Sleep | 1.0334275 | 0.34 | [0.36, 1.7] | 0.9209 | 1.0578502 | 0.36 | [0.36, 1.76] | 0.8680 |
| Anhedonia | ISI:Short Sleep | 1.0276950 | 0.01 | [1, 1.05] | 0.0169 | 1.0263169 | 0.01 | [1, 1.05] | 0.0269 |
| Anhedonia | ISI:Long Sleep | 1.0179864 | 0.03 | [0.97, 1.07] | 0.4791 | 1.0174797 | 0.03 | [0.97, 1.07] | 0.5023 |
| Depressed Mood | ISI | 1.0207236 | 0.01 | [1, 1.04] | 0.0243 | 1.0212582 | 0.01 | [1, 1.04] | 0.0244 |
| Depressed Mood | Short Sleep | 0.8370318 | 0.13 | [0.57, 1.1] | 0.2707 | 0.8319131 | 0.14 | [0.56, 1.1] | 0.2669 |
| Depressed Mood | Long Sleep | 1.5010144 | 0.48 | [0.56, 2.44] | 0.2049 | 1.5972814 | 0.53 | [0.57, 2.63] | 0.1553 |
| Depressed Mood | ISI:Short Sleep | 1.0022262 | 0.01 | [0.98, 1.02] | 0.8383 | 1.0027899 | 0.01 | [0.98, 1.02] | 0.8038 |
| Depressed Mood | ISI:Long Sleep | 0.9785476 | 0.02 | [0.93, 1.03] | 0.3841 | 0.9736618 | 0.02 | [0.92, 1.02] | 0.2992 |
| Appetite dysregulation | ISI | 1.0239255 | 0.01 | [1, 1.04] | 0.0203 | 1.0214125 | 0.01 | [1, 1.04] | 0.0400 |
| Appetite dysregulation | Short Sleep | 1.1797149 | 0.20 | [0.78, 1.58] | 0.3388 | 1.1016521 | 0.19 | [0.72, 1.48] | 0.5811 |
| Appetite dysregulation | Long Sleep | 1.9647520 | 0.68 | [0.63, 3.3] | 0.0529 | 1.8543470 | 0.65 | [0.59, 3.12] | 0.0773 |
| Appetite dysregulation | ISI:Short Sleep | 0.9935221 | 0.01 | [0.97, 1.02] | 0.5825 | 0.9968360 | 0.01 | [0.97, 1.02] | 0.7921 |
| Appetite dysregulation | ISI:Long Sleep | 0.9567349 | 0.03 | [0.9, 1.01] | 0.1111 | 0.9566688 | 0.03 | [0.9, 1.01] | 0.1121 |
| Fatigue | ISI | 1.0180811 | 0.01 | [1, 1.03] | 0.0103 | 1.0200161 | 0.01 | [1.01, 1.03] | 0.0051 |
| Fatigue | Short Sleep | 1.0142583 | 0.12 | [0.78, 1.25] | 0.9056 | 0.9865207 | 0.12 | [0.75, 1.22] | 0.9105 |
| Fatigue | Long Sleep | 0.9923559 | 0.26 | [0.49, 1.5] | 0.9765 | 0.9950127 | 0.26 | [0.49, 1.5] | 0.9846 |
| Fatigue | ISI:Short Sleep | 0.9995985 | 0.01 | [0.98, 1.02] | 0.9608 | 1.0022814 | 0.01 | [0.99, 1.02] | 0.7834 |
| Fatigue | ISI:Long Sleep | 1.0094111 | 0.02 | [0.97, 1.05] | 0.6328 | 1.0062077 | 0.02 | [0.97, 1.04] | 0.7520 |
| Difficulty concentrating | ISI | 1.0104140 | 0.01 | [0.99, 1.03] | 0.3482 | 1.0094975 | 0.01 | [0.99, 1.03] | 0.3993 |
| Difficulty concentrating | Short Sleep | 0.9278503 | 0.18 | [0.58, 1.27] | 0.6917 | 0.8792638 | 0.17 | [0.55, 1.21] | 0.5035 |
| Difficulty concentrating | Long Sleep | 1.2039904 | 0.50 | [0.23, 2.18] | 0.6522 | 1.0720115 | 0.44 | [0.21, 1.93] | 0.8656 |
| Difficulty concentrating | ISI:Short Sleep | 1.0048446 | 0.01 | [0.98, 1.03] | 0.7108 | 1.0095512 | 0.01 | [0.98, 1.04] | 0.4747 |
| Difficulty concentrating | ISI:Long Sleep | 0.9832799 | 0.03 | [0.92, 1.05] | 0.6023 | 0.9889700 | 0.03 | [0.93, 1.05] | 0.7312 |
| Feelings of failure | ISI | 1.0093809 | 0.01 | [0.99, 1.03] | 0.2936 | 1.0085071 | 0.01 | [0.99, 1.03] | 0.3495 |
| Feelings of failure | Short Sleep | 1.0274385 | 0.16 | [0.72, 1.33] | 0.8588 | 1.0298102 | 0.16 | [0.72, 1.34] | 0.8502 |
| Feelings of failure | Long Sleep | 1.3028895 | 0.40 | [0.52, 2.09] | 0.3896 | 1.3473157 | 0.42 | [0.52, 2.18] | 0.3440 |
| Feelings of failure | ISI:Short Sleep | 0.9960146 | 0.01 | [0.98, 1.02] | 0.7051 | 0.9955016 | 0.01 | [0.97, 1.02] | 0.6764 |
| Feelings of failure | ISI:Long Sleep | 0.9938423 | 0.02 | [0.95, 1.04] | 0.7957 | 0.9903489 | 0.02 | [0.94, 1.04] | 0.6930 |
| Psychomotor disturbance | ISI | 1.0336489 | 0.02 | [1, 1.07] | 0.0694 | 1.0212463 | 0.02 | [0.99, 1.06] | 0.2448 |
| Psychomotor disturbance | Short Sleep | 0.9574616 | 0.30 | [0.36, 1.55] | 0.8909 | 0.9233209 | 0.29 | [0.35, 1.5] | 0.8016 |
| Psychomotor disturbance | Long Sleep | 1.4870082 | 1.02 | [-0.52, 3.49] | 0.5650 | 1.3809087 | 0.95 | [-0.47, 3.23] | 0.6377 |
| Psychomotor disturbance | ISI:Short Sleep | 1.0035784 | 0.02 | [0.96, 1.05] | 0.8663 | 1.0108569 | 0.02 | [0.97, 1.05] | 0.6121 |
| Psychomotor disturbance | ISI:Long Sleep | 0.9674465 | 0.05 | [0.87, 1.07] | 0.5359 | 0.9825010 | 0.05 | [0.88, 1.08] | 0.7396 |
| Suicidal behavior | ISI | 1.0453295 | 0.02 | [1, 1.09] | 0.0540 | 1.0355107 | 0.02 | [0.99, 1.08] | 0.1286 |
| Suicidal behavior | Short Sleep | 1.2183004 | 0.49 | [0.25, 2.19] | 0.6262 | 1.3643363 | 0.55 | [0.28, 2.45] | 0.4428 |
| Suicidal behavior | Long Sleep | 1.0787691 | 0.89 | [-0.66, 2.82] | 0.9266 | 1.2844599 | 1.08 | [-0.83, 3.4] | 0.7656 |
| Suicidal behavior | ISI:Short Sleep | 0.9841912 | 0.03 | [0.93, 1.04] | 0.5538 | 0.9799659 | 0.03 | [0.93, 1.03] | 0.4527 |
| Suicidal behavior | ISI:Long Sleep | 1.0245926 | 0.06 | [0.91, 1.14] | 0.6807 | 1.0246740 | 0.06 | [0.9, 1.15] | 0.6866 |
The figure below shows how changing ISI score is related to adjusted PHQ-9 score by sleep duration in this group.
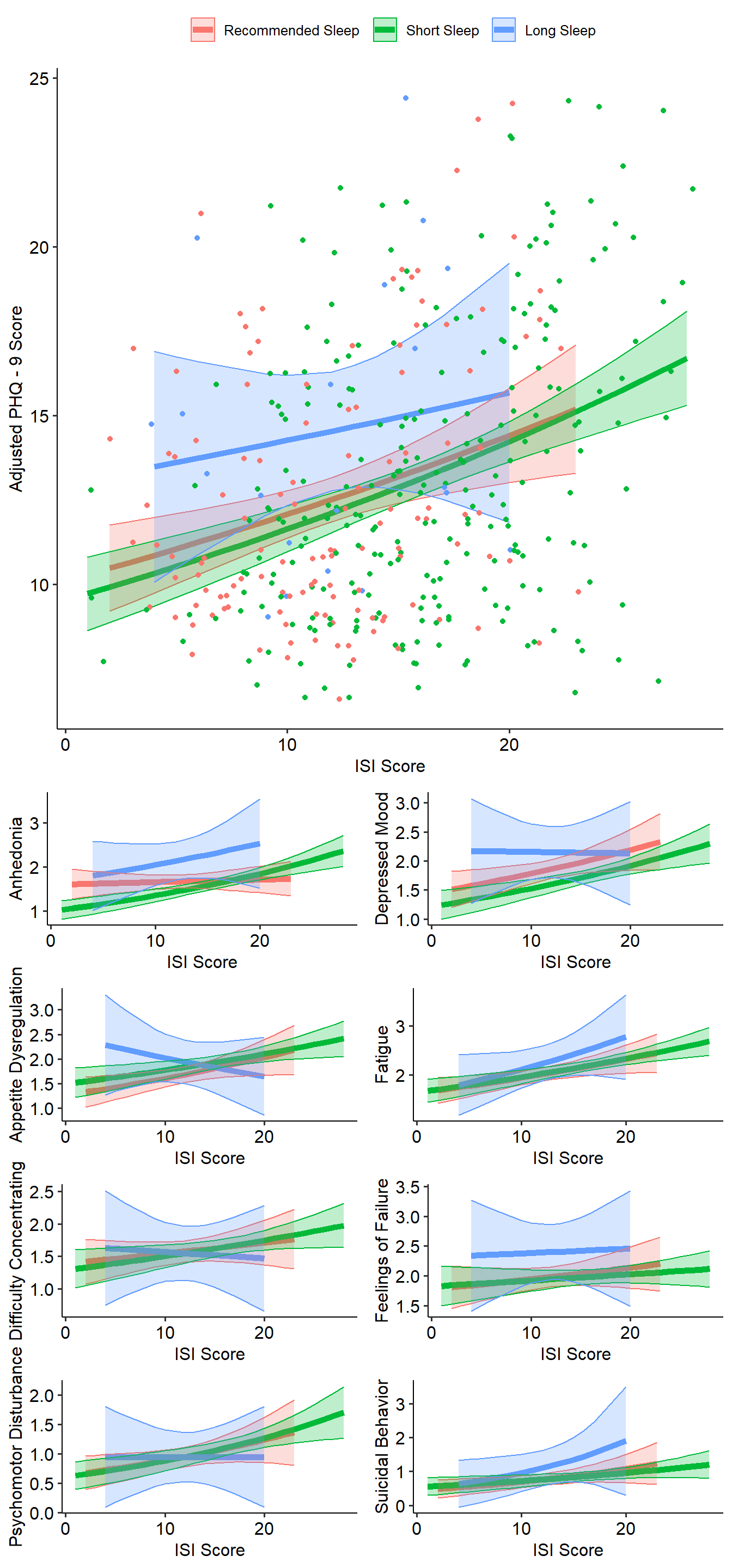
Model non-depressed subjects
Finally, let’s model the data for N=617 non-depressed individuals.
#### Analysis 3: Non-depressed Subjects Only ####
## Full Score
# Unadjusted
phq9.subset1 <- phq9.data[phq9.data$PHQtotal<10,]
phqscore.qp.sub1 <- glm(PHQtotal2~ISItotal*SleepDuration,family="quasipoisson",phq9.subset1)
phqscore.qp.sub1.maineff <- bind_cols("Symptom" = rep("Total",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(phqscore.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(phqscore.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(phqscore.qp.sub1)$coefficients[2:6,4])
# Adjusted for Age, Sex, Race, Education, and BMI
phqscore.qp.sub1.full <- glm(PHQtotal2~ISItotal*SleepDuration+Age+Sex+Race+Education+BMI,family="quasipoisson",phq9.subset1)
phqscore.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Total",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(phqscore.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(phqscore.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(phqscore.qp.sub1.full)$coefficients[c(2:4,16,17),4])
# Plot
phq9.subset1.predict <- predict(phqscore.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
phqscore.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
geom_point(aes(y = PHQtotal2),position="jitter")+
labs(x = "ISI Score",y="Adjusted PHQ - 9 Score")+
theme_pubr()+theme(legend.title = element_blank())
## Individual Symptoms
# Anhedonia
interest.qp.sub1 <- glm(PHQinterest~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
interest.qp.sub1.maineff <- bind_cols("Symptom" = rep("Interest",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(interest.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(interest.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(interest.qp.sub1)$coefficients[2:6,4])
interest.qp.sub1.full <- glm(PHQinterest~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
interest.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Interest",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(interest.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(interest.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(interest.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(interest.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
interest.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Anhedonia")+
theme_pubr()+theme(legend.position = "none")
# Depressed Mood
depress.qp.sub1 <- glm(PHQdepress~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
depress.qp.sub1.maineff <- bind_cols("Symptom" = rep("Depress",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(depress.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(depress.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(depress.qp.sub1)$coefficients[2:6,4])
depress.qp.sub1.full <- glm(PHQdepress~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
depress.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Depress",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(depress.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(depress.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(depress.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(depress.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
depress.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Depressed Mood")+
theme_pubr()+theme(legend.position = "none")
# Appetite
appetite.qp.sub1 <- glm(PHQappetite~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
appetite.qp.sub1.maineff <- bind_cols("Symptom" = rep("Appetite",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(appetite.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(appetite.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(appetite.qp.sub1)$coefficients[2:6,4])
appetite.qp.sub1.full <- glm(PHQappetite~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
appetite.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Appetite",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(appetite.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(appetite.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(appetite.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(appetite.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
appetite.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Appetite Dysregulation")+
theme_pubr()+theme(legend.position = "none")
# Fatigue
tired.qp.sub1 <- glm(PHQtired~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
tired.qp.sub1.maineff <- bind_cols("Symptom" = rep("Tired",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(tired.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(tired.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(tired.qp.sub1)$coefficients[2:6,4])
tired.qp.sub1.full <- glm(PHQtired~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
tired.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Tired",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(tired.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(tired.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(tired.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(tired.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
tired.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Fatigue")+
theme_pubr()+theme(legend.position = "none")
# Difficulty Concentrating
concentrate.qp.sub1 <- glm(PHQconcentrate~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
concentrate.qp.sub1.maineff <- bind_cols("Symptom" = rep("Concentrate",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(concentrate.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(concentrate.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(concentrate.qp.sub1)$coefficients[2:6,4])
concentrate.qp.sub1.full <- glm(PHQconcentrate~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
concentrate.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Concentrate",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(concentrate.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(concentrate.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(concentrate.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(concentrate.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
concentrate.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Difficulty Concentrating")+
theme_pubr()+theme(legend.position = "none")
# Feelings of Failure
failure.qp.sub1 <- glm(PHQfailure~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
failure.qp.sub1.maineff <- bind_cols("Symptom" = rep("Failure",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(failure.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(failure.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(failure.qp.sub1)$coefficients[2:6,4])
failure.qp.sub1.full <- glm(PHQfailure~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
failure.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Failure",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(failure.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(failure.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(failure.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(failure.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
failure.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Feelings of Failure")+
theme_pubr()+theme(legend.position = "none")
# Psychomotor Disturbance
psychomotor.qp.sub1 <- glm(PHQpsychomotor~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
psychomotor.qp.sub1.maineff <- bind_cols("Symptom" = rep("Psychomotor",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(psychomotor.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(psychomotor.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(psychomotor.qp.sub1)$coefficients[2:6,4])
psychomotor.qp.sub1.full <- glm(PHQpsychomotor~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
psychomotor.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Psychomotor",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(psychomotor.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(psychomotor.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(psychomotor.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(psychomotor.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
psychomotor.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Psychomotor Disturbance")+
theme_pubr()+theme(legend.position = "none")
# Suicidal behavior
suicide.qp.sub1 <- glm(PHQsuicide~ISItotal*SleepDuration,phq9.subset1,family="quasipoisson")
suicide.qp.sub1.maineff <- bind_cols("Symptom" = rep("Suicide",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(suicide.qp.sub1, "exp(b1)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub1, "exp(b2)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub1, "exp(b3)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub1, "exp(b4)", parameterNames=paste("b",0:5,sep="")),
deltaMethod(suicide.qp.sub1, "exp(b5)", parameterNames=paste("b",0:5,sep=""))) %>% as_tibble(),
"p-value" = summary(suicide.qp.sub1)$coefficients[2:6,4])
suicide.qp.sub1.full <- glm(PHQsuicide~ISItotal*SleepDuration+Age+Sex+Education+Race+BMI,phq9.subset1,family="quasipoisson")
suicide.qp.sub1.full.maineff <- bind_cols("Symptom" = rep("Suicide",5),
"Parameter" = c("ISI","Short Sleep","Long Sleep","ISI:Short Sleep","ISI:Long Sleep"),
bind_rows(deltaMethod(suicide.qp.sub1.full, "exp(b1)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub1.full, "exp(b2)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub1.full, "exp(b3)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub1.full, "exp(b15)", parameterNames=paste("b",0:16,sep="")),
deltaMethod(suicide.qp.sub1.full, "exp(b16)", parameterNames=paste("b",0:16,sep=""))) %>% as_tibble(),
"p-value" = summary(suicide.qp.sub1.full)$coefficients[c(2:4,16,17),4])
phq9.subset1.predict<- predict(suicide.qp.sub1, type="response",se.fit=TRUE)
phq9.subset1$fit <- as.numeric(phq9.subset1.predict$fit)
phq9.subset1$se.fit <- as.numeric(phq9.subset1.predict$se.fit)
suicide.sub1.plot <- ggplot(phq9.subset1, aes(x=ISItotal,y=fit,color=SleepDuration))+geom_line(size=2)+
geom_ribbon(aes(ymin=fit-1.96*se.fit,
ymax=fit+1.96*se.fit,fill=SleepDuration),alpha=.25)+
labs(x = "ISI Score",y="Suicidal Behavior")+
theme_pubr()+theme(legend.position = "none")
#### Table 4: Sleep influence by symptom in unadjusted/adjusted models for non-depressed only ####
phq9.symptom.sub1.summary <- bind_cols(
bind_rows(phqscore.qp.sub1.maineff,interest.qp.sub1.maineff,depress.qp.sub1.maineff,appetite.qp.sub1.maineff,tired.qp.sub1.maineff,
concentrate.qp.sub1.maineff,failure.qp.sub1.maineff,psychomotor.qp.sub1.maineff,suicide.qp.sub1.maineff) %>%
transmute(Symptom = factor(Symptom,levels = c("Total","Interest","Depress","Appetite","Tired","Concentrate","Failure","Psychomotor","Suicide"),
labels = c("Adjusted PHQ-9 Score","Anhedonia","Depressed Mood","Appetite dysregulation","Fatigue",
"Difficulty concentrating","Feelings of failure","Psychomotor disturbance","Suicidal behavior")),
Parameter,
"PR" = Estimate,
"SE" = round(SE,2),
"95% CI" = paste0("[",round(`2.5 %`,2),", ",round(`97.5 %`,2),"]"),
"p-value" = round(`p-value`,4)),
bind_rows(phqscore.qp.sub1.full.maineff,interest.qp.sub1.full.maineff,depress.qp.sub1.full.maineff,appetite.qp.sub1.full.maineff,
tired.qp.sub1.full.maineff,concentrate.qp.sub1.full.maineff,failure.qp.sub1.full.maineff,psychomotor.qp.sub1.full.maineff,
suicide.qp.sub1.full.maineff) %>%
transmute("aPR" = Estimate,
"aSE" = round(SE,2),
"a95% CI" = paste0("[",round(`2.5 %`,2),", ",round(`97.5 %`,2),"]"),
"ap-value" = round(`p-value`,4))
)
The table below shows their model results (as with the whole sample).
gt(phq9.symptom.sub1.summary)
| Symptom | Parameter | PR | SE | 95% CI | p-value | aPR | aSE | a95% CI | ap-value |
|---|---|---|---|---|---|---|---|---|---|
| Adjusted PHQ-9 Score | ISI | 1.0686013 | 0.01 | [1.05, 1.09] | 0.0000 | 1.0684075 | 0.01 | [1.05, 1.09] | 0.0000 |
| Adjusted PHQ-9 Score | Short Sleep | 1.4304891 | 0.17 | [1.09, 1.77] | 0.0034 | 1.4738678 | 0.18 | [1.11, 1.84] | 0.0020 |
| Adjusted PHQ-9 Score | Long Sleep | 0.8612373 | 0.23 | [0.42, 1.31] | 0.5721 | 0.8301561 | 0.22 | [0.4, 1.26] | 0.4827 |
| Adjusted PHQ-9 Score | ISI:Short Sleep | 0.9625754 | 0.01 | [0.94, 0.98] | 0.0006 | 0.9646551 | 0.01 | [0.94, 0.99] | 0.0020 |
| Adjusted PHQ-9 Score | ISI:Long Sleep | 1.0046695 | 0.02 | [0.96, 1.05] | 0.8424 | 1.0069011 | 0.02 | [0.96, 1.05] | 0.7691 |
| Anhedonia | ISI | 1.0510897 | 0.02 | [1.02, 1.08] | 0.0016 | 1.0550867 | 0.02 | [1.02, 1.09] | 0.0010 |
| Anhedonia | Short Sleep | 1.1877465 | 0.30 | [0.61, 1.77] | 0.4899 | 1.2250476 | 0.31 | [0.61, 1.84] | 0.4262 |
| Anhedonia | Long Sleep | 0.5938074 | 0.31 | [-0.02, 1.21] | 0.3248 | 0.6243789 | 0.33 | [-0.03, 1.27] | 0.3754 |
| Anhedonia | ISI:Short Sleep | 0.9620691 | 0.02 | [0.92, 1.01] | 0.0940 | 0.9582441 | 0.02 | [0.91, 1] | 0.0767 |
| Anhedonia | ISI:Long Sleep | 1.0495973 | 0.05 | [0.96, 1.14] | 0.2710 | 1.0455395 | 0.05 | [0.96, 1.14] | 0.3118 |
| Depressed Mood | ISI | 1.0477059 | 0.01 | [1.02, 1.07] | 0.0005 | 1.0497628 | 0.01 | [1.02, 1.08] | 0.0003 |
| Depressed Mood | Short Sleep | 1.3998214 | 0.28 | [0.85, 1.95] | 0.0925 | 1.4840074 | 0.30 | [0.89, 2.08] | 0.0542 |
| Depressed Mood | Long Sleep | 0.6269109 | 0.28 | [0.08, 1.17] | 0.2949 | 0.6079280 | 0.27 | [0.08, 1.14] | 0.2631 |
| Depressed Mood | ISI:Short Sleep | 0.9581394 | 0.02 | [0.92, 0.99] | 0.0241 | 0.9546943 | 0.02 | [0.92, 0.99] | 0.0189 |
| Depressed Mood | ISI:Long Sleep | 1.0372800 | 0.04 | [0.96, 1.12] | 0.3405 | 1.0396821 | 0.04 | [0.96, 1.12] | 0.3085 |
| Appetite dysregulation | ISI | 1.0609296 | 0.02 | [1.03, 1.09] | 0.0002 | 1.0587363 | 0.02 | [1.03, 1.09] | 0.0003 |
| Appetite dysregulation | Short Sleep | 1.7426111 | 0.38 | [1.01, 2.48] | 0.0102 | 1.8629647 | 0.41 | [1.06, 2.67] | 0.0050 |
| Appetite dysregulation | Long Sleep | 1.2526604 | 0.56 | [0.16, 2.34] | 0.6114 | 1.1705133 | 0.52 | [0.15, 2.19] | 0.7228 |
| Appetite dysregulation | ISI:Short Sleep | 0.9675740 | 0.02 | [0.93, 1.01] | 0.1030 | 0.9653307 | 0.02 | [0.93, 1.01] | 0.0941 |
| Appetite dysregulation | ISI:Long Sleep | 0.9835712 | 0.04 | [0.9, 1.07] | 0.6977 | 0.9926187 | 0.04 | [0.91, 1.08] | 0.8616 |
| Fatigue | ISI | 1.0857724 | 0.01 | [1.07, 1.1] | 0.0000 | 1.0832091 | 0.01 | [1.06, 1.1] | 0.0000 |
| Fatigue | Short Sleep | 1.4311582 | 0.20 | [1.05, 1.82] | 0.0093 | 1.4279642 | 0.20 | [1.03, 1.82] | 0.0117 |
| Fatigue | Long Sleep | 1.0100373 | 0.30 | [0.43, 1.59] | 0.9730 | 0.9640999 | 0.28 | [0.41, 1.52] | 0.9011 |
| Fatigue | ISI:Short Sleep | 0.9610082 | 0.01 | [0.94, 0.98] | 0.0010 | 0.9697321 | 0.01 | [0.95, 0.99] | 0.0152 |
| Fatigue | ISI:Long Sleep | 0.9841725 | 0.03 | [0.93, 1.04] | 0.5455 | 0.9908502 | 0.03 | [0.94, 1.04] | 0.7261 |
| Difficulty concentrating | ISI | 1.0848619 | 0.02 | [1.05, 1.12] | 0.0000 | 1.0828414 | 0.02 | [1.05, 1.12] | 0.0000 |
| Difficulty concentrating | Short Sleep | 1.6476865 | 0.42 | [0.82, 2.48] | 0.0524 | 1.6833236 | 0.45 | [0.8, 2.56] | 0.0512 |
| Difficulty concentrating | Long Sleep | 0.6408899 | 0.41 | [-0.17, 1.45] | 0.4888 | 0.5712258 | 0.38 | [-0.17, 1.31] | 0.3951 |
| Difficulty concentrating | ISI:Short Sleep | 0.9430012 | 0.02 | [0.9, 0.99] | 0.0113 | 0.9515255 | 0.02 | [0.91, 1] | 0.0430 |
| Difficulty concentrating | ISI:Long Sleep | 1.0030407 | 0.05 | [0.9, 1.11] | 0.9556 | 1.0050883 | 0.06 | [0.9, 1.11] | 0.9275 |
| Feelings of failure | ISI | 1.0394952 | 0.02 | [1.01, 1.07] | 0.0165 | 1.0430680 | 0.02 | [1.01, 1.08] | 0.0114 |
| Feelings of failure | Short Sleep | 1.2784439 | 0.30 | [0.69, 1.86] | 0.2920 | 1.3399599 | 0.32 | [0.7, 1.98] | 0.2273 |
| Feelings of failure | Long Sleep | 0.8019526 | 0.40 | [0.02, 1.58] | 0.6557 | 0.7949801 | 0.40 | [0.01, 1.58] | 0.6470 |
| Feelings of failure | ISI:Short Sleep | 0.9800119 | 0.02 | [0.94, 1.02] | 0.3594 | 0.9805971 | 0.02 | [0.94, 1.03] | 0.3994 |
| Feelings of failure | ISI:Long Sleep | 1.0212248 | 0.05 | [0.93, 1.11] | 0.6447 | 1.0228449 | 0.05 | [0.93, 1.11] | 0.6226 |
| Psychomotor disturbance | ISI | 1.1307922 | 0.03 | [1.07, 1.19] | 0.0000 | 1.1319852 | 0.03 | [1.07, 1.19] | 0.0000 |
| Psychomotor disturbance | Short Sleep | 1.3340265 | 0.57 | [0.21, 2.46] | 0.5020 | 1.2246660 | 0.55 | [0.15, 2.3] | 0.6520 |
| Psychomotor disturbance | Long Sleep | 0.6218032 | 0.68 | [-0.71, 1.96] | 0.6644 | 0.5269802 | 0.61 | [-0.67, 1.72] | 0.5805 |
| Psychomotor disturbance | ISI:Short Sleep | 0.9553425 | 0.03 | [0.89, 1.02] | 0.1887 | 0.9584913 | 0.04 | [0.89, 1.03] | 0.2581 |
| Psychomotor disturbance | ISI:Long Sleep | 1.0061248 | 0.08 | [0.84, 1.17] | 0.9414 | 1.0046670 | 0.09 | [0.83, 1.18] | 0.9578 |
| Suicidal behavior | ISI | 1.1327105 | 0.05 | [1.03, 1.23] | 0.0060 | 1.1298284 | 0.05 | [1.03, 1.23] | 0.0090 |
| Suicidal behavior | Short Sleep | 2.1137730 | 1.49 | [-0.8, 5.03] | 0.2884 | 2.7774162 | 2.02 | [-1.19, 6.74] | 0.1612 |
| Suicidal behavior | Long Sleep | 3.8123764 | 4.29 | [-4.6, 12.23] | 0.2352 | 4.1331270 | 4.57 | [-4.83, 13.1] | 0.2001 |
| Suicidal behavior | ISI:Short Sleep | 0.9323474 | 0.06 | [0.82, 1.04] | 0.2376 | 0.9101974 | 0.06 | [0.79, 1.03] | 0.1502 |
| Suicidal behavior | ISI:Long Sleep | 0.8988875 | 0.10 | [0.7, 1.1] | 0.3489 | 0.8853279 | 0.10 | [0.69, 1.08] | 0.2709 |
The figure below shows how changing ISI score is related to adjusted PHQ-9 score by sleep duration in this group.